The smile is related or not?
Karim Mezhoud
2019-05-28
library(data.table)
library(dplyr)
library(plyr)
library(stringr)
library(EBImage)
library(keras)sample_submission <- fread("sample_submission.csv")
head(sample_submission)## img_pair is_related
## 1: face05508.jpg-face01210.jpg 0
## 2: face05750.jpg-face00898.jpg 0
## 3: face05820.jpg-face03938.jpg 0
## 4: face02104.jpg-face01172.jpg 0
## 5: face02428.jpg-face05611.jpg 0
## 6: face01219.jpg-face00274.jpg 0relationship <- data.table::fread("train_relationships.csv",stringsAsFactors = TRUE)
relationship %>%
filter(p1 == "F0058/MID2")## p1 p2
## 1 F0058/MID2 F0058/MID4
## 2 F0058/MID2 F0058/MID5
## 3 F0058/MID2 F0058/MID11 Processing relationships
The goal is to group and list the folders with kinship.
1.1 Collapse from p1 to p2
collaps_P1P2 <- plyr::ddply(relationship,c("p1"),
function(df1)paste(df1$p2,
collapse = ","))
colnames(collaps_P1P2) <- c("p1", "p2")
collaps_P1P2 %>%
head(20) ## p1 p2
## 1 F0002/MID1 F0002/MID3
## 2 F0002/MID2 F0002/MID3
## 3 F0005/MID1 F0005/MID2
## 4 F0005/MID3 F0005/MID2
## 5 F0009/MID1 F0009/MID4,F0009/MID3,F0009/MID2,F0009/MID6
## 6 F0009/MID2 F0009/MID4,F0009/MID6,F0009/MID3
## 7 F0009/MID3 F0009/MID4,F0009/MID6
## 8 F0009/MID4 F0009/MID6
## 9 F0009/MID5 F0009/MID1,F0009/MID2,F0009/MID4,F0009/MID6,F0009/MID3
## 10 F0009/MID7 F0009/MID3,F0009/MID6,F0009/MID4,F0009/MID1,F0009/MID2
## 11 F0010/MID1 F0010/MID3,F0010/MID4
## 12 F0010/MID2 F0010/MID3,F0010/MID4
## 13 F0010/MID3 F0010/MID4
## 14 F0016/MID1 F0016/MID5,F0016/MID2
## 15 F0016/MID2 F0016/MID4,F0016/MID3
## 16 F0016/MID3 F0016/MID4
## 17 F0017/MID1 F0017/MID3,F0017/MID4,F0017/MID7,F0017/MID5,F0017/MID6
## 18 F0017/MID2 F0017/MID5,F0017/MID4,F0017/MID3,F0017/MID7,F0017/MID6
## 19 F0017/MID3 F0017/MID4,F0017/MID6,F0017/MID5
## 20 F0017/MID4 F0017/MID5,F0017/MID61.2 Cancatenate subfamilies 1
subfamilies1 <- paste(collaps_P1P2$p1, collaps_P1P2$p2, sep=",")
## extract subfamilies with more than two persons
subfamilies1_ <- subfamilies1[stringr::str_detect(subfamilies1, "\\w\\d*\\/\\w*\\d*,\\w\\d*\\/\\w*\\d*")]
subfamilies1_ %>%
head(20)## [1] "F0002/MID1,F0002/MID3"
## [2] "F0002/MID2,F0002/MID3"
## [3] "F0005/MID1,F0005/MID2"
## [4] "F0005/MID3,F0005/MID2"
## [5] "F0009/MID1,F0009/MID4,F0009/MID3,F0009/MID2,F0009/MID6"
## [6] "F0009/MID2,F0009/MID4,F0009/MID6,F0009/MID3"
## [7] "F0009/MID3,F0009/MID4,F0009/MID6"
## [8] "F0009/MID4,F0009/MID6"
## [9] "F0009/MID5,F0009/MID1,F0009/MID2,F0009/MID4,F0009/MID6,F0009/MID3"
## [10] "F0009/MID7,F0009/MID3,F0009/MID6,F0009/MID4,F0009/MID1,F0009/MID2"
## [11] "F0010/MID1,F0010/MID3,F0010/MID4"
## [12] "F0010/MID2,F0010/MID3,F0010/MID4"
## [13] "F0010/MID3,F0010/MID4"
## [14] "F0016/MID1,F0016/MID5,F0016/MID2"
## [15] "F0016/MID2,F0016/MID4,F0016/MID3"
## [16] "F0016/MID3,F0016/MID4"
## [17] "F0017/MID1,F0017/MID3,F0017/MID4,F0017/MID7,F0017/MID5,F0017/MID6"
## [18] "F0017/MID2,F0017/MID5,F0017/MID4,F0017/MID3,F0017/MID7,F0017/MID6"
## [19] "F0017/MID3,F0017/MID4,F0017/MID6,F0017/MID5"
## [20] "F0017/MID4,F0017/MID5,F0017/MID6"1.3 Collapse from p2 to p1
collaps_P2P1 <- plyr::ddply(relationship,c("p2"),
function(df1)paste(df1$p1,
collapse = ","))
colnames(collaps_P2P1) <- c("p2", "p1")
collaps_P2P1 %>%
head(20)## p2
## 1 F0002/MID3
## 2 F0005/MID2
## 3 F0009/MID1
## 4 F0009/MID2
## 5 F0009/MID3
## 6 F0009/MID4
## 7 F0009/MID6
## 8 F0010/MID3
## 9 F0010/MID4
## 10 F0016/MID2
## 11 F0016/MID3
## 12 F0016/MID4
## 13 F0016/MID5
## 14 F0017/MID3
## 15 F0017/MID4
## 16 F0017/MID5
## 17 F0017/MID6
## 18 F0017/MID7
## 19 F0020/MID1
## 20 F0020/MID10
## p1
## 1 F0002/MID1,F0002/MID2
## 2 F0005/MID1,F0005/MID3
## 3 F0009/MID5,F0009/MID7
## 4 F0009/MID1,F0009/MID5,F0009/MID7
## 5 F0009/MID1,F0009/MID2,F0009/MID5,F0009/MID7
## 6 F0009/MID1,F0009/MID2,F0009/MID3,F0009/MID5,F0009/MID7
## 7 F0009/MID1,F0009/MID2,F0009/MID3,F0009/MID4,F0009/MID5,F0009/MID7
## 8 F0010/MID1,F0010/MID2
## 9 F0010/MID1,F0010/MID2,F0010/MID3
## 10 F0016/MID1
## 11 F0016/MID2
## 12 F0016/MID2,F0016/MID3
## 13 F0016/MID1
## 14 F0017/MID1,F0017/MID2
## 15 F0017/MID1,F0017/MID2,F0017/MID3
## 16 F0017/MID1,F0017/MID2,F0017/MID3,F0017/MID4
## 17 F0017/MID1,F0017/MID2,F0017/MID3,F0017/MID4
## 18 F0017/MID1,F0017/MID2,F0017/MID5
## 19 F0020/MID7
## 20 F0020/MID1,F0020/MID11,F0020/MID2,F0020/MID3,F0020/MID5,F0020/MID6,F0020/MID7,F0020/MID81.4 Cancatenate subfamilies 2
subfamilies2 <- paste(collaps_P2P1$p1, collaps_P2P1$p2, sep=",")
## extract subfamilies with more than two persons
subfamilies2_ <- subfamilies2[stringr::str_detect(subfamilies2, "\\w\\d*\\/\\w*\\d*,\\w\\d*\\/\\w*\\d*")]
subfamilies2_ %>%
head(20)## [1] "F0002/MID1,F0002/MID2,F0002/MID3"
## [2] "F0005/MID1,F0005/MID3,F0005/MID2"
## [3] "F0009/MID5,F0009/MID7,F0009/MID1"
## [4] "F0009/MID1,F0009/MID5,F0009/MID7,F0009/MID2"
## [5] "F0009/MID1,F0009/MID2,F0009/MID5,F0009/MID7,F0009/MID3"
## [6] "F0009/MID1,F0009/MID2,F0009/MID3,F0009/MID5,F0009/MID7,F0009/MID4"
## [7] "F0009/MID1,F0009/MID2,F0009/MID3,F0009/MID4,F0009/MID5,F0009/MID7,F0009/MID6"
## [8] "F0010/MID1,F0010/MID2,F0010/MID3"
## [9] "F0010/MID1,F0010/MID2,F0010/MID3,F0010/MID4"
## [10] "F0016/MID1,F0016/MID2"
## [11] "F0016/MID2,F0016/MID3"
## [12] "F0016/MID2,F0016/MID3,F0016/MID4"
## [13] "F0016/MID1,F0016/MID5"
## [14] "F0017/MID1,F0017/MID2,F0017/MID3"
## [15] "F0017/MID1,F0017/MID2,F0017/MID3,F0017/MID4"
## [16] "F0017/MID1,F0017/MID2,F0017/MID3,F0017/MID4,F0017/MID5"
## [17] "F0017/MID1,F0017/MID2,F0017/MID3,F0017/MID4,F0017/MID6"
## [18] "F0017/MID1,F0017/MID2,F0017/MID5,F0017/MID7"
## [19] "F0020/MID7,F0020/MID1"
## [20] "F0020/MID1,F0020/MID11,F0020/MID2,F0020/MID3,F0020/MID5,F0020/MID6,F0020/MID7,F0020/MID8,F0020/MID10"families <- unique(c(subfamilies1_, subfamilies2_))
length(families)## [1] 30231.5 Index of person in the same family
family_idx <-
unique(unlist(strsplit(families, ","))) %>%
str_extract("\\w\\d*") %>%
as.factor() %>%
as.numeric()
family_idx## [1] 1 1 1 2 2 2 3 3 3 3 3 3 3 4 4 4 4
## [18] 5 5 5 5 5 6 6 6 6 6 6 6 7 7 7 7 7
## [35] 7 7 7 7 7 8 8 8 8 8 9 9 9 9 9 9 10
## [52] 10 10 10 10 11 11 11 11 12 12 12 12 12 13 13 13 13
## [69] 13 13 13 14 14 14 14 14 14 15 15 15 16 16 17 17 17
## [86] 17 17 17 18 18 19 19 19 19 19 20 20 20 21 21 21 21
## [103] 21 21 22 22 22 22 23 23 23 23 24 24 24 24 24 24 25
## [120] 25 25 25 26 26 27 27 27 27 27 27 28 28 28 28 28 29
## [137] 29 29 29 29 29 29 30 30 30 30 30 30 31 31 31 31 31
## [154] 32 32 32 32 32 32 32 32 33 33 33 33 34 34 34 34 34
## [171] 34 34 34 35 35 35 35 35 36 36 36 36 37 37 37 37 37
## [188] 37 37 37 37 37 38 38 38 38 38 38 38 38 38 38 38 38
## [205] 38 38 38 39 39 39 39 39 39 40 40 40 40 40 40 40 41
## [222] 41 41 41 42 42 42 42 43 43 43 43 44 44 44 44 44 45
## [239] 45 45 45 45 45 45 46 46 46 47 47 47 47 47 47 48 48
## [256] 48 48 48 48 48 49 49 49 50 50 50 50 50 50 51 51 51
## [273] 51 51 51 51 51 52 52 52 52 52 52 52 52 53 53 53 53
## [290] 53 54 54 55 55 55 56 56 56 57 57 58 58 58 58 59 59
## [307] 59 59 60 60 60 60 60 60 60 60 61 61 61 61 61 61 61
## [324] 61 62 62 62 63 63 63 63 64 64 64 64 64 65 65 65 65
## [341] 66 66 66 66 66 66 66 66 67 67 67 68 68 68 68 68 69
## [358] 69 69 69 70 70 70 70 70 70 71 71 71 71 72 72 72 72
## [375] 72 72 72 72 72 73 73 73 73 73 73 73 73 74 74 74 74
## [392] 75 75 75 75 76 76 76 76 77 77 77 77 77 77 77 78 78
## [409] 78 79 79 79 79 80 80 80 80 80 81 81 81 81 81 81 82
## [426] 82 82 82 83 83 83 83 83 83 83 83 84 84 84 84 84 84
## [443] 85 85 85 86 86 86 86 87 87 87 87 87 87 88 88 88 88
## [460] 88 89 89 89 89 89 89 90 90 90 90 90 91 91 91 91 91
## [477] 91 91 91 91 91 92 92 92 92 92 92 92 92 92 92 92 92
## [494] 92 92 93 93 93 93 93 93 93 94 94 94 94 95 95 95 95
## [511] 96 96 96 96 96 97 97 97 97 98 98 98 98 98 98 98 98
## [528] 99 99 99 99 99 99 99 100 100 100 100 101 101 101 101 101 102
## [545] 102 102 102 103 103 103 103 103 103 103 103 103 103 103 103 103 103
## [562] 104 104 104 104 104 105 105 105 105 106 106 106 106 106 106 107 107
## [579] 107 107 107 107 107 108 108 108 108 109 109 109 109 110 110 110 110
## [596] 111 111 111 111 111 112 112 112 113 113 113 113 113 113 113 113 113
## [613] 113 114 114 114 114 115 115 115 116 116 116 116 117 117 118 118 118
## [630] 118 118 119 119 119 120 120 120 120 120 120 120 121 121 121 121 121
## [647] 121 121 121 121 122 122 123 123 124 124 125 125 126 126 126 126 127
## [664] 127 127 127 128 128 128 128 128 128 128 128 128 128 129 129 129 129
## [681] 129 130 130 130 130 130 130 131 131 131 131 131 131 132 132 132 132
## [698] 133 133 133 133 133 133 134 134 134 134 134 135 135 135 135 136 136
## [715] 136 136 136 136 137 137 137 137 137 138 138 138 138 139 139 139 139
## [732] 139 139 139 139 139 139 139 140 140 140 140 140 140 141 141 142 142
## [749] 142 142 143 143 143 144 144 144 145 145 146 146 146 146 146 146 146
## [766] 146 147 147 147 147 147 147 148 148 148 148 148 148 149 149 150 150
## [783] 150 150 150 151 151 151 152 152 152 152 153 153 153 153 153 154 154
## [800] 154 154 154 154 154 154 154 154 154 154 154 155 155 155 155 156 156
## [817] 156 156 156 156 157 157 157 157 158 158 158 158 158 158 158 158 159
## [834] 159 159 160 160 160 160 161 161 161 161 161 161 161 162 162 162 163
## [851] 163 163 163 164 164 164 164 164 164 164 164 164 164 165 165 165 165
## [868] 165 165 166 166 166 167 167 167 167 167 167 167 168 168 168 168 168
## [885] 168 168 168 168 168 168 169 169 169 169 169 169 170 170 170 170 171
## [902] 171 171 172 172 172 172 172 172 172 172 173 173 173 173 173 174 174
## [919] 174 174 175 175 175 175 176 176 177 177 177 178 178 178 178 178 178
## [936] 179 179 179 180 180 180 180 180 181 181 181 181 181 181 181 181 181
## [953] 181 182 182 182 182 182 182 182 183 183 183 184 184 184 184 184 184
## [970] 184 184 184 184 185 185 185 185 185 186 186 186 186 186 186 187 187
## [987] 188 188 188 188 188 188 189 189 189 189 189 190 190 190 190 190 190
## [1004] 190 191 191 191 191 191 191 191 191 191 192 192 192 192 193 193 193
## [1021] 193 193 194 194 194 194 195 195 195 195 195 195 195 196 196 196 197
## [1038] 197 197 197 198 198 198 198 198 198 198 198 199 199 199 199 199 199
## [1055] 199 199 199 199 200 200 200 201 201 201 202 202 202 202 203 203 203
## [1072] 204 204 204 205 205 205 205 205 206 206 206 206 206 206 206 207 207
## [1089] 207 207 207 207 207 207 208 208 208 209 209 209 209 209 209 210 210
## [1106] 210 211 211 211 211 212 212 212 212 213 213 213 213 214 214 214 214
## [1123] 214 215 215 215 216 216 216 216 216 217 217 217 218 218 218 218 218
## [1140] 219 219 219 219 219 220 220 221 221 221 221 221 221 221 221 221 221
## [1157] 221 222 222 223 223 223 223 223 224 224 224 224 225 225 225 225 226
## [1174] 226 226 226 227 227 227 227 227 228 228 229 229 229 230 230 230 230
## [1191] 230 231 231 231 231 232 232 232 232 233 233 233 233 234 234 234 234
## [1208] 234 234 235 235 235 235 235 235 236 236 236 236 236 236 237 237 237
## [1225] 237 237 238 238 238 238 239 239 239 239 240 240 240 240 240 241 241
## [1242] 241 241 241 241 242 242 242 242 243 243 243 243 243 243 243 243 243
## [1259] 243 243 243 243 244 244 244 244 244 245 245 245 245 246 246 246 246
## [1276] 247 247 247 247 247 248 248 248 248 249 249 249 249 249 249 249 249
## [1293] 249 250 250 250 251 251 251 251 252 252 252 252 252 252 253 253 253
## [1310] 254 254 254 254 254 254 254 254 255 255 255 256 256 256 256 256 257
## [1327] 257 257 257 257 258 258 258 258 258 258 259 259 259 259 260 260 260
## [1344] 260 260 260 261 261 261 261 261 261 261 261 261 261 261 261 261 261
## [1361] 261 261 261 261 261 261 261 261 261 261 261 261 261 261 261 261 261
## [1378] 261 261 261 261 261 261 261 261 261 261 261 262 262 262 263 263 263
## [1395] 263 264 264 264 265 265 265 265 265 265 265 266 266 266 266 266 266
## [1412] 266 267 267 267 267 267 268 268 269 269 269 269 269 269 270 270 270
## [1429] 271 271 271 272 272 272 272 272 273 273 273 273 273 274 274 274 274
## [1446] 274 274 274 275 275 275 275 275 275 275 276 276 276 276 276 276 276
## [1463] 277 277 277 277 277 277 277 278 278 278 278 278 279 279 279 279 279
## [1480] 279 280 280 280 280 281 281 281 282 282 282 282 283 283 283 283 283
## [1497] 283 283 283 284 284 285 285 285 285 286 286 286 286 286 286 286 286
## [1514] 286 287 287 287 287 287 288 288 288 288 289 289 289 290 290 290 290
## [1531] 290 290 290 291 291 291 291 291 291 291 292 292 292 292 292 293 293
## [1548] 293 293 293 293 294 294 295 295 295 295 295 296 296 297 297 297 297
## [1565] 297 298 298 298 298 298 298 299 299 299 299 299 300 300 300 301 301
## [1582] 301 301 302 302 302 302 303 303 303 303 303 303 303 303 303 303 304
## [1599] 304 304 305 305 305 305 306 306 306 306 306 307 307 307 307 307 307
## [1616] 307 308 308 308 308 308 308 308 308 308 309 309 309 309 310 310 310
## [1633] 310 311 311 311 311 311 311 311 311 311 311 312 312 312 312 313 313
## [1650] 313 313 314 314 314 314 314 315 315 315 315 315 315 315 316 316 316
## [1667] 316 317 317 317 317 317 317 318 318 318 319 319 319 320 320 320 320
## [1684] 321 321 321 321 321 322 322 322 322 322 322 323 323 323 323 323 324
## [1701] 324 324 324 324 325 325 325 325 326 326 326 326 326 326 326 326 326
## [1718] 327 327 327 327 327 328 328 328 328 328 328 329 329 329 329 329 329
## [1735] 329 329 330 330 330 330 331 331 331 331 332 332 332 332 333 333 334
## [1752] 334 334 334 334 334 334 334 334 334 335 335 335 335 335 335 335 335
## [1769] 336 336 336 336 336 337 337 338 338 338 338 339 339 339 340 340 340
## [1786] 340 340 340 341 341 341 341 341 341 341 341 341 342 342 342 342 342
## [1803] 343 343 343 343 343 343 344 344 344 344 344 344 344 345 345 345 345
## [1820] 346 346 346 346 346 346 347 347 347 347 347 347 348 348 348 348 348
## [1837] 348 348 349 349 349 350 350 350 351 351 351 351 351 352 352 352 353
## [1854] 353 353 353 353 353 353 353 353 353 354 354 354 354 355 355 355 355
## [1871] 355 356 356 357 357 357 357 357 357 357 357 358 358 358 358 358 358
## [1888] 358 359 359 359 359 359 360 360 361 361 361 361 361 362 362 362 362
## [1905] 363 363 363 363 363 363 363 363 364 364 364 364 364 364 365 365 366
## [1922] 366 366 366 366 366 367 367 367 368 368 368 368 368 368 368 368 369
## [1939] 369 369 370 370 370 370 371 371 371 371 371 372 372 372 372 372 372
## [1956] 372 372 372 373 373 373 373 373 373 374 374 374 374 374 374 374 374
## [1973] 375 375 375 375 376 376 376 376 376 376 376 376 376 376 376 377 377
## [1990] 377 377 378 378 378 378 378 379 379 379 379 379 380 380 380 380 380
## [2007] 380 380 381 381 381 381 381 382 382 382 382 382 382 383 383 383 383
## [2024] 383 383 383 383 383 383 384 384 384 384 384 385 385 386 386 387 387
## [2041] 387 387 387 387 388 388 388 388 389 389 389 389 389 390 390 390 391
## [2058] 391 391 391 392 392 392 392 392 392 393 393 394 394 394 395 395 395
## [2075] 395 395 395 396 396 397 397 398 398 398 399 399 400 400 400 401 401
## [2092] 402 402 402 402 403 403 403 403 404 404 405 405 405 406 406 407 407
## [2109] 407 407 407 407 407 407 408 408 408 408 408 409 409 409 409 409 409
## [2126] 410 410 410 410 410 410 411 411 411 411 411 412 412 412 413 413 413
## [2143] 413 414 414 414 414 415 415 415 415 416 416 416 417 417 417 417 417
## [2160] 418 418 418 418 418 418 418 419 419 419 420 420 420 421 421 421 421
## [2177] 421 422 422 422 422 423 423 424 424 424 424 424 425 425 425 425 426
## [2194] 426 426 426 427 427 427 427 428 428 428 428 428 428 429 429 429 430
## [2211] 430 430 430 430 430 430 430 431 431 431 431 431 432 432 432 432 432
## [2228] 433 433 433 433 433 433 434 434 434 434 435 435 435 435 435 436 436
## [2245] 436 436 437 437 437 437 438 438 438 438 439 439 439 440 440 440 441
## [2262] 441 441 442 442 442 443 443 443 443 444 444 444 444 445 445 445 445
## [2279] 445 445 445 445 445 446 446 446 446 446 447 447 447 447 447 447 448
## [2296] 448 448 448 448 448 448 448 449 449 449 449 450 450 450 450 451 451
## [2313] 451 451 452 452 452 452 453 453 453 454 454 454 454 455 455 456 456
## [2330] 456 456 457 457 457 457 457 457 458 458 458 458 458 458 459 459 459
## [2347] 459 459 459 459 459 460 460 460 460 460 461 461 461 462 462 462 462
## [2364] 463 463 463 463 463 463 463 464 464 464 464 465 465 465 465 465 465
## [2381] 465 465 465 466 466 466 466 466 467 467 467 467 467 468 468 468 468
## [2398] 469 469 469 469 469 469 470 470 470 470 470 470 470 470 4701.6 Collapse Persons by family index
fam <- as.data.frame(cbind(Person = unique(unlist(strsplit(families, ","))),family_idx))
fam %>%
head(20)## Person family_idx
## 1 F0002/MID1 1
## 2 F0002/MID3 1
## 3 F0002/MID2 1
## 4 F0005/MID1 2
## 5 F0005/MID2 2
## 6 F0005/MID3 2
## 7 F0009/MID1 3
## 8 F0009/MID4 3
## 9 F0009/MID3 3
## 10 F0009/MID2 3
## 11 F0009/MID6 3
## 12 F0009/MID5 3
## 13 F0009/MID7 3
## 14 F0010/MID1 4
## 15 F0010/MID3 4
## 16 F0010/MID4 4
## 17 F0010/MID2 4
## 18 F0016/MID1 5
## 19 F0016/MID5 5
## 20 F0016/MID2 5unique_families <- plyr::ddply(fam,c("family_idx"),
function(df1)paste(df1$Person,
collapse = ","))
colnames(unique_families) <- c("index", "Persons with kinship relationships")
DT::datatable(unique_families) %>%
DT::formatStyle( colnames(unique_families), color = 'black')There are 470 kinship relationships
unique_families$`Persons with kinship relationships`[1:10]## [1] "F0002/MID1,F0002/MID3,F0002/MID2"
## [2] "F0024/MID2,F0024/MID1,F0024/MID3,F0024/MID5,F0024/MID4"
## [3] "F0225/MID1,F0225/MID2,F0225/MID3,F0225/MID4"
## [4] "F0227/MID1,F0227/MID3,F0227/MID5,F0227/MID2,F0227/MID4"
## [5] "F0228/MID1,F0228/MID3,F0228/MID4,F0228/MID2"
## [6] "F0231/MID1,F0231/MID7,F0231/MID11,F0231/MID10,F0231/MID12,F0231/MID13,F0231/MID14,F0231/MID2,F0231/MID5,F0231/MID4,F0231/MID6,F0231/MID9,F0231/MID3,F0231/MID8"
## [7] "F0233/MID1,F0233/MID5,F0233/MID3,F0233/MID2,F0233/MID6"
## [8] "F0236/MID1,F0236/MID4,F0236/MID3,F0236/MID2"
## [9] "F0237/MID1,F0237/MID3,F0237/MID2,F0237/MID4,F0237/MID5,F0237/MID6"
## [10] "F0238/MID1,F0238/MID4,F0238/MID8,F0238/MID3,F0238/MID7,F0238/MID5,F0238/MID6"#length(str_split(unique_families$`Persons with kinship relationships`[10],pattern = ","))3 Check folder and files availabilities
The goal is to clean up is_related_all and no_related_all from not existing folders or images.
is_related_all <-
is_related_all %>%
filter(file.exists(paste0("train/", p1))) %>%
filter(file.exists(paste0("train/", p2))) %>%
filter(!length(list.files(paste0("train/", p1))) == 0) %>%
filter(!length(list.files(paste0("train/", p2))) == 0)
no_related_all <-
no_related_all %>%
filter(file.exists(paste0("train/", p1))) %>%
filter(file.exists(paste0("train/", p2))) %>%
filter(!(length(list.files(paste0("train/", p1))) == 0)) %>%
filter(!(length(list.files(paste0("train/", p2))) == 0))
is_related_all %>%
filter(p1 == "F0460/MID8") ## p1 p2 is_related
## 1 F0460/MID8 F0460/MID1 1
## 2 F0460/MID8 F0460/MID5 1
## 3 F0460/MID8 F0460/MID6 1
## 4 F0460/MID8 F0460/MID7 1length(list.files("train/F0460/MID8/"))## [1] 0Note filter can not detect particular empty folders
kinships_all <-
is_related_all %>%
bind_rows(no_related_all) %>%
arrange(by = p1)
kinships_all %>%
head(8)## p1 p2 is_related
## 1 F0002/MID1 F0002/MID3 1
## 2 F0002/MID1 F0002/MID2 0
## 3 F0002/MID2 F0002/MID3 1
## 4 F0002/MID2 F0002/MID1 0
## 5 F0002/MID3 F0002/MID1 1
## 6 F0002/MID3 F0002/MID2 1
## 7 F0024/MID1 F0024/MID2 1
## 8 F0024/MID1 F0024/MID3 14 Images Data processing
6 Keras tentative
6.1 sample test and Train data
set.seed(982)
idx <- seq_len(nrow(trainx1))
idx_train <- sample(idx, max(idx) * 0.8, replace = FALSE)
train_x1 <- trainx1[idx_train,]
test_x1 <- trainx1[-idx_train,]
train_x2 <- trainx2[idx_train,]
test_x2 <- trainx2[-idx_train,]
Labels_train <- keras::to_categorical(Labels[idx_train])
Labels_test <- keras::to_categorical(Labels[idx[-idx_train]])
dim(train_x1)## [1] 9979 2352str(Labels_test)## num [1:2495, 1:2] 0 0 1 0 1 1 1 1 0 1 ...6.2 Set the model
# input1 <- layer_input(shape = c(28, 28, 3))
# input2 <- layer_input(shape = c(28, 28, 3))
#
# conv1 <- input1 %>%
# layer_conv_2d(filters = 7, kernel_size = c(1, 1), padding='same', activation='relu') %>%
# layer_conv_2d(filters = 7, kernel_size = c(3, 3), padding='same', activation='relu')
#
# conv2 <- input2 %>%
# layer_conv_2d(filters = 7, kernel_size = c(1, 1), padding='same', activation='relu') %>%
# layer_conv_2d(filters = 7, kernel_size = c(3, 3), padding='same', activation='relu')
#
#
# output <-
# layer_concatenate(c(conv1, conv2), axis = 1) %>%
# layer_dense(units = 256, activation = 'relu', input_shape = c(2352)) %>% # 28 x 28 x 3
# layer_dense(units = 128, activation = 'relu') %>%
# layer_dense(units = 2, activation = 'softmax') # we have two output 0,1
input1 <- layer_input(shape = c(2352))
input2 <- layer_input(shape = c(2352))
#conv1 <- layer_conv_2d( filters = 4, input_shape = c(28, 28, 3), kernel_size = 3)
#conv2 <- layer_conv_2d( filters = 4, input_shape = c(28, 28, 3), kernel_size = 3)
output <-
layer_concatenate(list(input1, input2)) %>%
# layer_dense(units = 512, activation = 'relu',
# kernel_regularizer = regularizer_l2(l = 0.001)) %>%
# layer_dropout(rate = 0.1) %>%
# layer_dense(units = 256, activation = 'relu',
# kernel_regularizer = regularizer_l2(l = 0.001)) %>%
# layer_dropout(rate = 0.05) %>%
layer_dense(units = 256, activation = 'relu',
kernel_regularizer = regularizer_l2(l = 0.001)) %>%
# layer_dropout(rate = 0.02) %>%
layer_dense(units = 64, activation = 'relu',
kernel_regularizer = regularizer_l2(l = 0.001)) %>%
# layer_dropout(rate = 0.01) %>%
layer_dense(units = 2, activation = 'softmax') # we have two output 0,1
#layer_flatten() %>%
#layer_dense(units = 2, activation = "softmax")
model <-
keras_model(inputs = list(input1, input2), outputs = output)
model %>% compile(optimizer = "adam", # , optimizer_rmsprop()
loss = 'binary_crossentropy',
metrics = 'accuracy')
summary(model)## ___________________________________________________________________________
## Layer (type) Output Shape Param # Connected to
## ===========================================================================
## input_1 (InputLayer) (None, 2352) 0
## ___________________________________________________________________________
## input_2 (InputLayer) (None, 2352) 0
## ___________________________________________________________________________
## concatenate (Concatenat (None, 4704) 0 input_1[0][0]
## input_2[0][0]
## ___________________________________________________________________________
## dense (Dense) (None, 256) 1204480 concatenate[0][0]
## ___________________________________________________________________________
## dense_1 (Dense) (None, 64) 16448 dense[0][0]
## ___________________________________________________________________________
## dense_2 (Dense) (None, 2) 130 dense_1[0][0]
## ===========================================================================
## Total params: 1,221,058
## Trainable params: 1,221,058
## Non-trainable params: 0
## ___________________________________________________________________________6.3 train the model
history <-
model %>%
fit(list(train_x1, train_x2),
Labels_train,
epochs = 100,
batch_size = 256,
verbose = 1, # 2
validation_split = 0.2) # list(test_x1, Labels_test),
plot(history)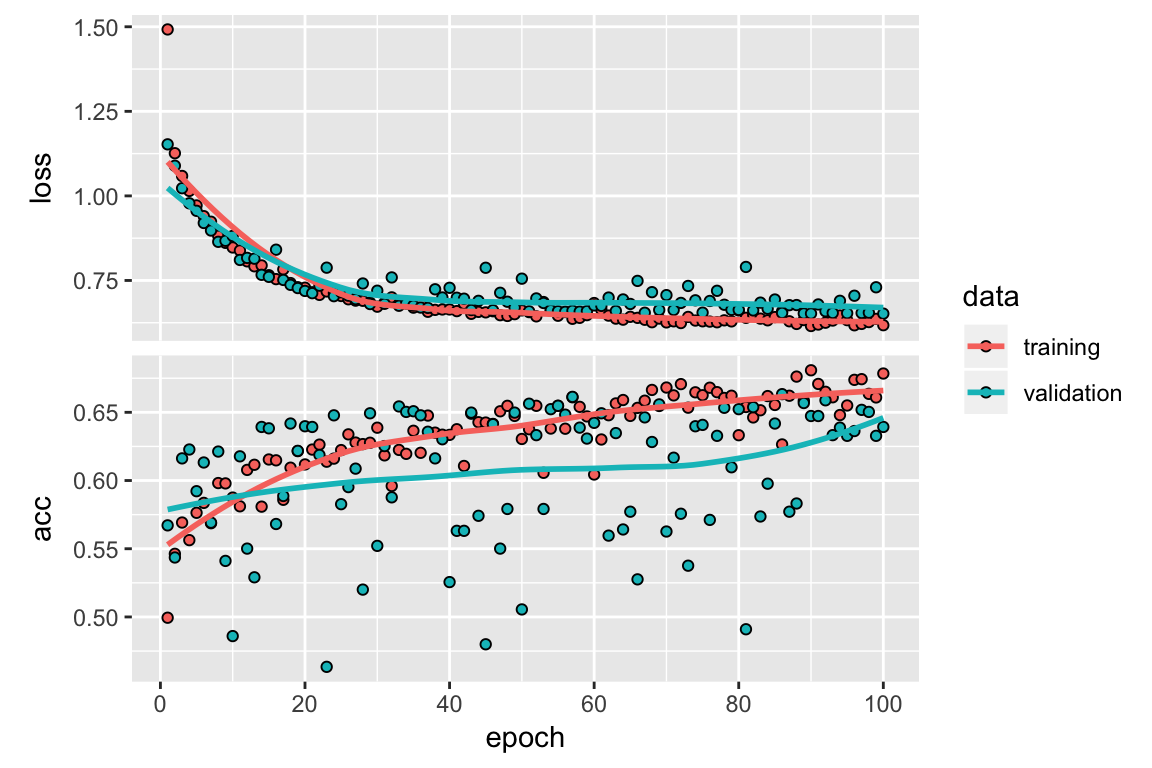
6.4 Evaluate
score <- model %>% evaluate(
list(test_x1, test_x2),
Labels_test,
verbose = 0
)
# Output metrics
cat('Test loss:', score[[1]], '\n')## Test loss: 0.6601205cat('Test accuracy:', score[[2]], '\n')## Test accuracy: 0.62645297 Prediction
sample_submission <- fread("sample_submission.csv")
head(sample_submission)## img_pair is_related
## 1: face05508.jpg-face01210.jpg 0
## 2: face05750.jpg-face00898.jpg 0
## 3: face05820.jpg-face03938.jpg 0
## 4: face02104.jpg-face01172.jpg 0
## 5: face02428.jpg-face05611.jpg 0
## 6: face01219.jpg-face00274.jpg 07.1 get testx1 and testx2 from sample_submission file
sample_submission[, c("p1", "p2") := tstrsplit(img_pair, "-", fixed=TRUE)]
sample_submission %>%
head()## img_pair is_related p1 p2
## 1: face05508.jpg-face01210.jpg 0 face05508.jpg face01210.jpg
## 2: face05750.jpg-face00898.jpg 0 face05750.jpg face00898.jpg
## 3: face05820.jpg-face03938.jpg 0 face05820.jpg face03938.jpg
## 4: face02104.jpg-face01172.jpg 0 face02104.jpg face01172.jpg
## 5: face02428.jpg-face05611.jpg 0 face02428.jpg face05611.jpg
## 6: face01219.jpg-face00274.jpg 0 face01219.jpg face00274.jpgget_test_matrix <- function(df, col){
pathfile <- paste0("test/", df[col])
## Check if the path exists && the folder is not empty
if(file.exists(pathfile)){
# load image
img <- EBImage::readImage(pathfile)
# resize images
img <- EBImage::resize(img, 28, 28)
## reshape images
img <- keras::array_reshape(img, c(28,28,3))
testx <- rbind(testx, img)
}else{
to_delete <<- c(to_delete, paste0(pathfile))
}
return(testx)
}
to_delete <- NULL
testx <- NULL
star <- Sys.time()
testx1 <- apply(sample_submission, 1, function(row) get_test_matrix(row, 3))
end <- Sys.time()
print(end - star)## Time difference of 1.266156 minstestx1 <- t(testx1)
str(testx1)## num [1:5310, 1:2352] 0.481 0.146 0.455 0.647 0.655 ...to_delete## NULLto_delete <- NULL
testx <- NULL
star <- Sys.time()
testx2 <- apply(sample_submission, 1, function(row) get_test_matrix(row, 4))
end <- Sys.time()
print(end - star)## Time difference of 1.360726 minstestx2 <- t(testx2)
str(testx2)## num [1:5310, 1:2352] 0.431 0.315 0.383 0.517 0.751 ...to_delete## NULLis_related <- predict(model, list(testx1, testx2))
is_related %>%
head()## [,1] [,2]
## [1,] 0.4591229 0.5408770
## [2,] 0.6044239 0.3955760
## [3,] 0.5666160 0.4333840
## [4,] 0.6910377 0.3089623
## [5,] 0.6339806 0.3660194
## [6,] 0.5956860 0.40431408 submission
dt_submission <- data.table(
img_pair = sample_submission$img_pair,
is_related = is_related[,2])
head(dt_submission)## img_pair is_related
## 1: face05508.jpg-face01210.jpg 0.5408770
## 2: face05750.jpg-face00898.jpg 0.3955760
## 3: face05820.jpg-face03938.jpg 0.4333840
## 4: face02104.jpg-face01172.jpg 0.3089623
## 5: face02428.jpg-face05611.jpg 0.3660194
## 6: face01219.jpg-face00274.jpg 0.4043140fwrite(dt_submission, file = "submission.csv")9 save model
#model %>% save_model_hdf5("my_model.h5")
#new_model <- load_model_hdf5("my_model.h5")